Pneumonia Patient Condition Classification Using Diffusion Models and CLIP
Published:
Project Overview
Pneumonia remains a leading cause of childhood mortality. This project explores the application of diffusion models and contrastive language–image pre-training (CLIP) for pediatric chest X-ray classification into normal, viral pneumonia, and bacterial pneumonia categories.
We used a dataset of 5,856 images from Guangzhou Women and Children’s Medical Center. To address class imbalance, we fine-tuned a Stable Diffusion model using LoRA, generating 2,000 synthetic images for minority classes. We then fine-tuned a CLIP ViT-L/14 model to classify the combined dataset using prompt-based text labels.
Key Methods
- Stable Diffusion v2 + LoRA for image synthesis.
- CLIP fine-tuning for contrastive learning.
- Prompt engineering: “An image of [Type] chest X-ray.”
- Low GPU budget: only ~1% of parameters trained.
Results
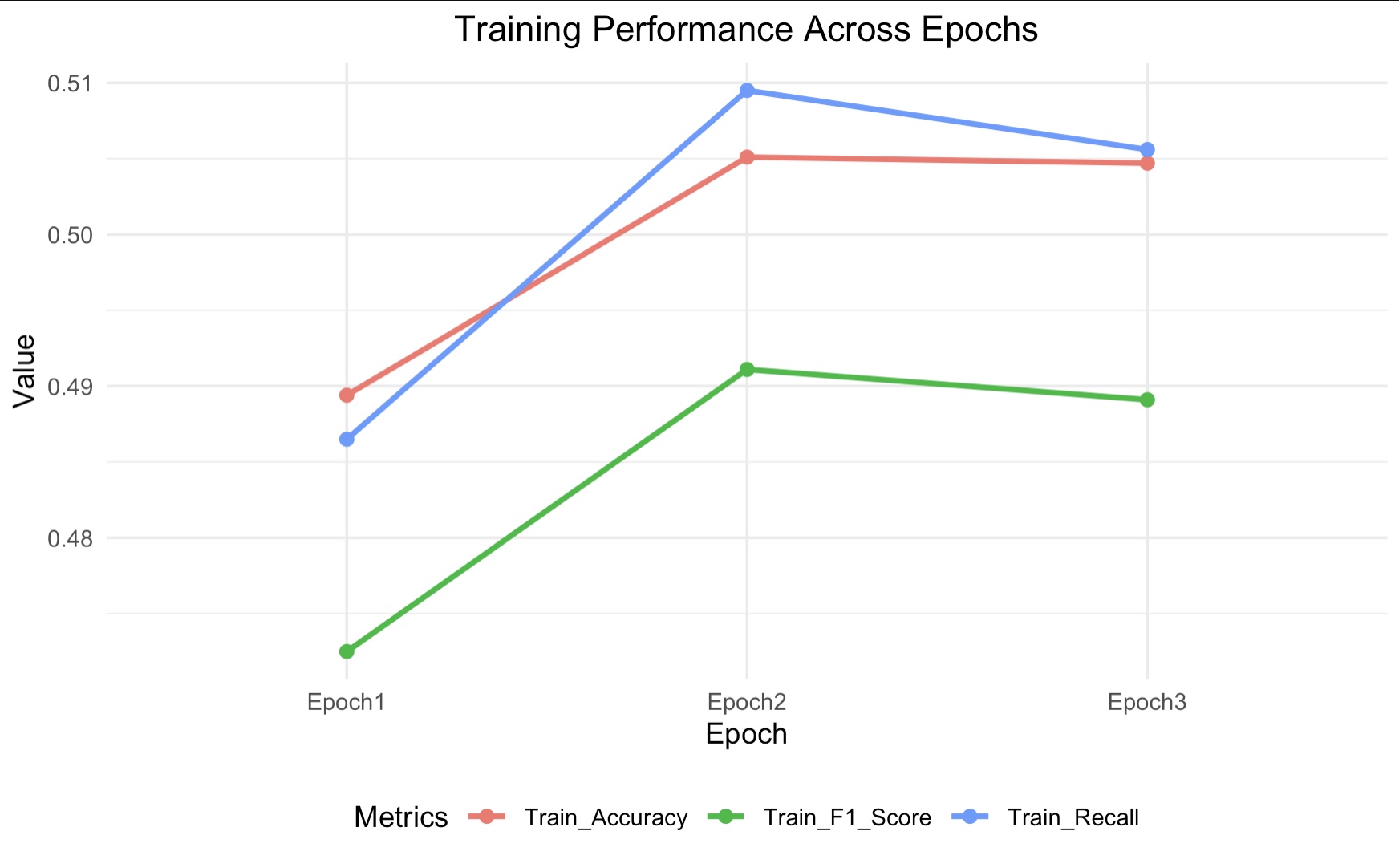
- Training accuracy increased from 48.94% to 50.51%
- Test accuracy: 37.5%, Recall: 50%
- Visual summary of metrics across epochs:
Tools & Environment
- Python 3.9, PyTorch 2.5.1
- NVIDIA RTX 3090 (24GB)
- LoRA, HuggingFace 🤗, OpenCLIP
Contributions
- Xiaomeng Xu: Code editing; abstract; introduction
- Wenfei Mao: Code editing; Diffusion Model; CLIP; Conclusion
- Yingzhen Wang: Code editing; Results; Diffusion model
- Shuoyuan Gao: Code editing; Experiment Setup; Conclusion
- Full GitHub Repo: View on GitHub
Future Work
- Try faster diffusion models (e.g., Consistency Models, One-Step Diffusion)
- Explore RL-based fine-tuning (e.g., DPO, PPO)
- Move from LoRA to full model fine-tuning for higher fidelity.
Summary
This study demonstrates a hybrid approach using generative models for class balancing and multimodal LLMs for medical image classification — showing promise despite current accuracy limitations.